Identifying Errors
By default, GRR displays sib pairs, half sibs, parent-offspring pairs, and
unrelated pairs. The user can select at any time which pairings are to be
displayed, and the category of other relative pairs is by default
deselected. As can be seen from our sample dataset, displayed
below, there are several errors in this pedigree file.
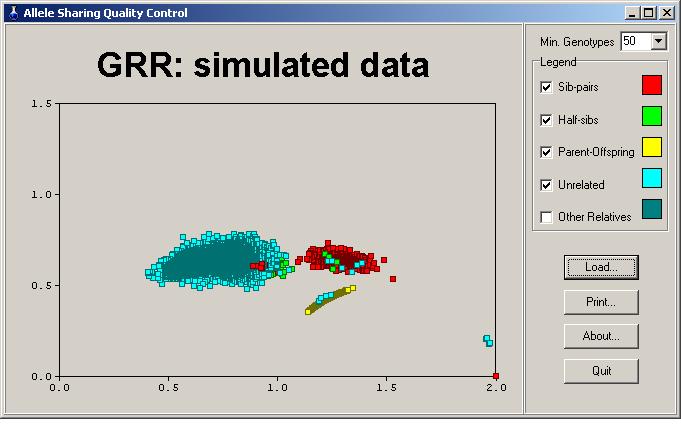
Sib pairs who are really MZ twins
We have a red dot in the bottom left corner, a coded sib pair whose mean IBS
is about 2; that is, they are virtually identical at every locus, indicative of
being MZ twins. Now click on that datapoint. A popup box appears
showing that in fact there are two sib pairs at that position, one pair in
family 28 and another in family 63.
Duplicated families
Note the appearance of cyan coloured squares near the MZ pair datapoints.
These are likely a duplicated family in the dataset. Note that their mean
IBS is very near 2, but not quite. Perhaps these individuals were
genotyped twice and now we have an estimate of the genotype error rate in this
study. When clicking on these points, we see a perfect correspondence
between family 155 and 175, with the same ID numbers used, supporting our
theory. Upon examining the cyan squares in the parent-offspring and
sib-pair clusters, the same family numbers appear.
Full-sibs who are really half-sibs
Now we deselect the unrelated pairs. We see a green cluster of
half-sibs with some red sib-pair dots in it. Those are pairs thought to be
full sibs, but are likely half-sibs. There appear to be six such
misclassified pairs.
Half-sibs who are really full-sibs
We see within the red cluster of full sibs several green dots; these
individuals are coded as half sibs, but are almost certainly
full-sibs. There appear to be four such pairs in our sample.
In the next section we examine other options
available in GRR.
| 


