PEDSTATS Tutorial - Input Files
PEDSTATS provides graphical and text summaries of the information contained in any pair of pedigree and data files.
Pedigree (.ped) files describe relationships between individuals in your dataset and also store marker genotypes,
disease status and quantitative trait values. Data (.dat) files provide a description of the contents of the
associated pedigree file.
PEDSTATS supports input files in either QTDT, LINKAGE or MENDEL
format . Although the three formats are similar, in the discussion below we will focus on QTDT format.
Describing Relationships Between Individuals
Although pedigrees can become quite complex, all the information that is necessary to
reconstruct individual relationships in a pedigree file can be summarized in five items:
a family identifier, an individual identifier, a link to each parent (if available) and
finally an indicator of each individual's sex.
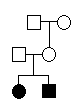 As an example of how family relationships are described, we will construct a pedigree
file for a small pedigree with two siblings, their parents and maternal grand-parents.
As an example of how family relationships are described, we will construct a pedigree
file for a small pedigree with two siblings, their parents and maternal grand-parents.
For this simple pedigree, the five key items take the following values:
FAMILY PERSON FATHER MOTHER SEX
example granpa unknown unknown m
example granny unknown unknown f
example father unknown unknown m
example mother granpa granny f
example sister father mother f
example brother father mother m
These key values constitute the first five columns of any pedigree
file. Because of restrictions in early genetic programs, text identifiers
are usually replaced by unique numeric values. After replacing each
identifier with unique integer and recoding sexes as 2 (female) and 1 (male),
this is what a basic space-delimited pedigree file would look like:
<contents of basic.ped>
1 1 0 0 1
1 2 0 0 2
1 3 0 0 1
1 4 1 2 2
1 5 3 4 2
1 6 3 4 1
<end of basic.ped>
A pedigree file can include multiple families. Each family can
have a unique structure, independent of other families in the dataset.
Describing Phenotypes and Genotypes
Usually the five standard columns are followed by various
types of genetic data, including phenotypes for discrete and quantitative
traits and marker genotypes.
Disease status is usually encoded in a single column as
U or 1 for unaffecteds,
A or 2 for affecteds, and
X or 0 for missing phenotypes.
Quantitative traits are encoded as numeric values with X
denoting missing values (it is also possible to use a peculiar numeric
value to flag missing phenotypes, but the procedure is prone to error
and not recommended).
Marker genotypes are encoded as two consecutive integers,
one for each allele, optionally separated by a "/". A 0 (zero) or X
can be used as a placeholder for missing alleles. The following are all
valid genotype entries 1/1 (homozygote for allele 1), 0/0
(missing genotype), and 3 4 (heterozygote for alleles 3 and 4). For
the X chromosome, males should be encoded as if they had two identical
alleles.
This is what the previous pedigree file might look like after adding a
column for disease status, measurements for a quantitative trait and
genotypes for two markers:
<contents of basic2.ped>
1 1 0 0 1 1 x 3 3 x x
1 2 0 0 2 1 x 4 4 x x
1 3 0 0 1 1 x 1 2 x x
1 4 1 2 2 1 x 4 3 x x
1 5 3 4 2 2 1.234 1 3 2 2
1 6 3 4 1 2 4.321 2 4 2 2
<end of basic2.ped>
Notice that the two siblings (individuals 5 and 6 in the last two rows)
are marked as affected (value 2 in the sixth column), everyone else is marked
as unaffected (value 1 in the sixth column). The
quantitative trait (seventh column) takes values 1.234 and 4.321 for individuals 5 and 6 respectively. Whereas
everyone is genotyped at the first marker, for the second marker, only
individuals 5 and 6 are genotyped.
Describing the pedigree file
Pedigree files can include any number of marker genotype, disease
status and quantitative trait variables, limited only by available
memory. Since each pedigree file has a unique structure (apart from
the first five columns), its contents must be described in a companion
data file.
The data file includes one row per data item in the pedigree file,
indicating the data type (encoded as M - marker, A - affection status,
T - Quantitative Trait C - Covariate and Z - Twins ) and providing a
one-word label
for each item. A data file for the pedigree above, which has one affection
status, followed by one quantitative trait and two marker genotypes might
read:
<contents of basic2.dat>
A some_disease
T some_trait
M some_marker
M another_marker
<end of basic2.dat>
Now that you understand pedigree and data file formats, you'll probably want to actually run PEDSTATS! You can
get a copy from our download page or if you'd like, you can take a look
at some text or PDF output first.
| 


