SamFile Class Reference
Allows the user to easily read/write a SAM/BAM file. More...
#include <SamFile.h>
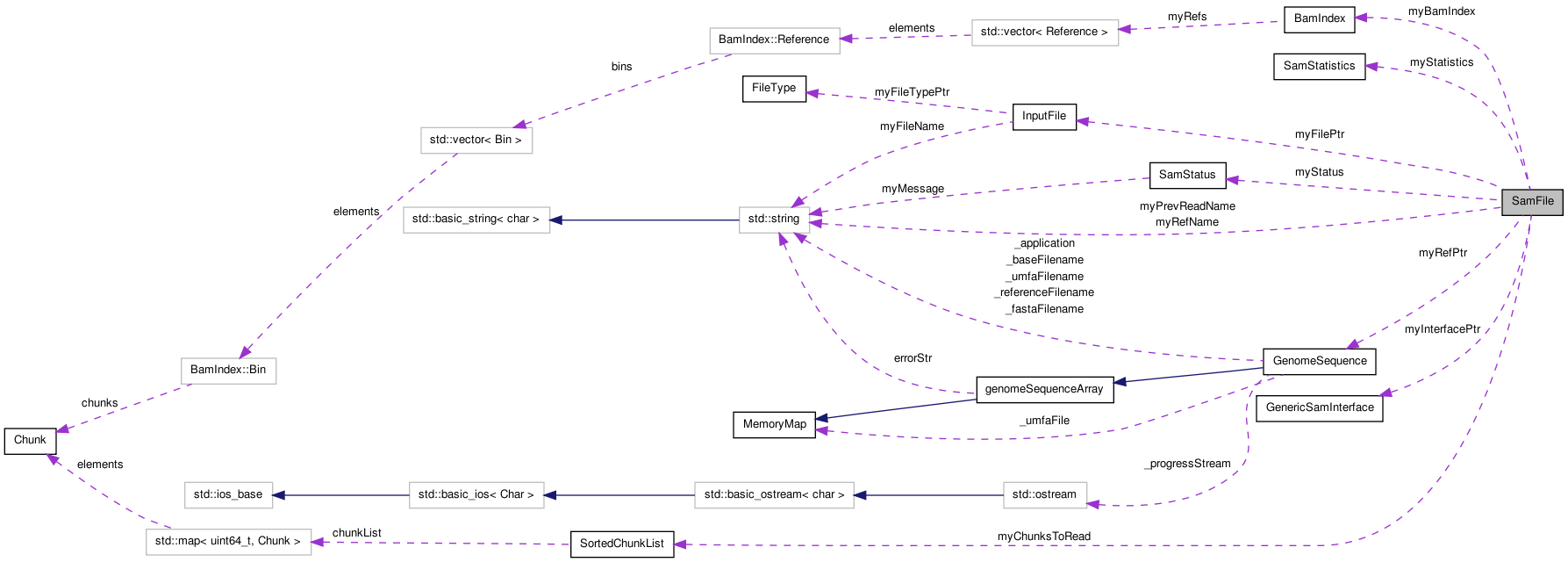
Public Types | |
| enum | OpenType { READ, WRITE } |
Enum for indicating whether to open the file for read or write. More... | |
| enum | SortedType { UNSORTED = 0, FLAG, COORDINATE, QUERY_NAME } |
Enum for indicating the type of sort for the file. More... | |
Public Member Functions | |
| SamFile () | |
| Default Constructor. | |
| SamFile (ErrorHandler::HandlingType errorHandlingType) | |
| Constructor that sets the error handling type. | |
| SamFile (const char *filename, OpenType mode) | |
| Constructor that opens the specified file based on the specified mode (READ/WRITE). | |
| SamFile (const char *filename, OpenType mode, ErrorHandler::HandlingType errorHandlingType) | |
| Constructor that opens the specified file based on the specified mode (READ/WRITE) and handles errors per the specified handleType. | |
| SamFile (const char *filename, OpenType mode, SamFileHeader *header) | |
| Constructor that opens the specified file based on the specified mode (READ/WRITE). | |
| SamFile (const char *filename, OpenType mode, ErrorHandler::HandlingType errorHandlingType, SamFileHeader *header) | |
| Constructor that opens the specified file based on the specified mode (READ/WRITE) and handles errors per the specified handleType. | |
| bool | OpenForRead (const char *filename, SamFileHeader *header=NULL) |
| Open a sam/bam file for reading with the specified filename. | |
| bool | OpenForWrite (const char *filename, SamFileHeader *header=NULL) |
| Open a sam/bam file for writing with the specified filename. | |
| bool | ReadBamIndex (const char *filename) |
| Read the specified bam index file. | |
| bool | ReadBamIndex () |
| Read the bam index file using the BAM filename as a base. | |
| void | SetReference (GenomeSequence *reference) |
| Sets the reference to the specified genome sequence object. | |
| void | SetReadSequenceTranslation (SamRecord::SequenceTranslation translation) |
| Set the type of sequence translation to use when reading the sequence. | |
| void | SetWriteSequenceTranslation (SamRecord::SequenceTranslation translation) |
| Set the type of sequence translation to use when writing the sequence. | |
| void | Close () |
| Close the file if there is one open. | |
| bool | IsOpen () |
| Returns whether or not the file has been opened successfully. | |
| bool | IsEOF () |
| Returns whether or not the end of the file has been reached. | |
| bool | ReadHeader (SamFileHeader &header) |
| Reads the header section from the file and stores it in the passed in header. | |
| bool | WriteHeader (SamFileHeader &header) |
| Writes the specified header into the file. | |
| bool | ReadRecord (SamFileHeader &header, SamRecord &record) |
| Reads the next record from the file & stores it in the passed in record. | |
| bool | WriteRecord (SamFileHeader &header, SamRecord &record) |
| Writes the specified record into the file. | |
| void | setSortedValidation (SortedType sortType) |
| Set the flag to validate that the file is sorted as it is read/written. | |
| uint32_t | GetCurrentRecordCount () |
| Return the number of records that have been read/written so far. | |
| SamStatus::Status | GetFailure () |
| Get the Status of the last call that sets status. | |
| SamStatus::Status | GetStatus () |
| Get the Status of the last call that sets status. | |
| const char * | GetStatusMessage () |
| Get the Status of the last call that sets status. | |
| bool | SetReadSection (int32_t refID) |
| Sets what part of the BAM file should be read. | |
| bool | SetReadSection (const char *refName) |
| Sets what part of the BAM file should be read. | |
| bool | SetReadSection (int32_t refID, int32_t start, int32_t end, bool overlap=true) |
| Sets what part of the BAM file should be read. | |
| bool | SetReadSection (const char *refName, int32_t start, int32_t end, bool overlap=true) |
| Sets what part of the BAM file should be read. | |
| int32_t | getNumMappedReadsFromIndex (int32_t refID) |
| Get the number of mapped reads in the specified reference id. | |
| int32_t | getNumUnMappedReadsFromIndex (int32_t refID) |
| Get the number of unmapped reads in the specified reference id. | |
| int32_t | getNumMappedReadsFromIndex (const char *refName, SamFileHeader &header) |
| Get the number of mapped reads in the specified reference name. | |
| int32_t | getNumUnMappedReadsFromIndex (const char *refName, SamFileHeader &header) |
| Get the number of unmapped reads in the specified reference name. | |
| uint32_t | GetNumOverlaps (SamRecord &samRecord) |
| Returns the number of bases in the passed in read that overlap the region that is currently set. | |
| void | GenerateStatistics (bool genStats) |
| Whether or not statistics should be generated for this file. | |
| const BamIndex * | GetBamIndex () |
| Return the bam index if one has been opened. | |
| long int | GetCurrentPosition () |
| Get the current file position. | |
| void | DisableBuffering () |
| void | PrintStatistics () |
| bool | attemptRecoverySync (bool(*checkSignature)(void *data), int length) |
| void | setAttemptRecovery (bool flag=false) |
Protected Member Functions | |
| void | init (const char *filename, OpenType mode, SamFileHeader *header) |
| void | resetFile () |
| Resets the file prepping for a new file. | |
| bool | validateSortOrder (SamRecord &record, SamFileHeader &header) |
| Validate that the record is sorted compared to the previously read record if there is one, according to the specified sort order. | |
| SortedType | getSortOrderFromHeader (SamFileHeader &header) |
| bool | readIndexedRecord (SamFileHeader &header, SamRecord &record) |
| Overwrites read record to read from the specific reference only. | |
| bool | processNewSection (SamFileHeader &header) |
Protected Attributes | |
| IFILE | myFilePtr |
| GenericSamInterface * | myInterfacePtr |
| bool | myIsOpenForRead |
| Flag to indicate if a file is open for reading. | |
| bool | myIsOpenForWrite |
| Flag to indicate if a file is open for writing. | |
| bool | myHasHeader |
| Flag to indicate if a header has been read/written - required before being able to read/write a record. | |
| SortedType | mySortedType |
| int32_t | myPrevCoord |
| Previous values used for checking if the file is sorted. | |
| int32_t | myPrevRefID |
| std::string | myPrevReadName |
| uint32_t | myRecordCount |
| Keep a count of the number of records that have been read/written so far. | |
| SamStatistics * | myStatistics |
| Pointer to the statistics for this file. | |
| SamStatus | myStatus |
| The status of the last SamFile command. | |
| bool | myIsBamOpenForRead |
| Values for reading Sorted BAM files via the index. | |
| bool | myNewSection |
| bool | myOverlapSection |
| int32_t | myRefID |
| int32_t | myStartPos |
| int32_t | myEndPos |
| uint64_t | myCurrentChunkEnd |
| SortedChunkList | myChunksToRead |
| BamIndex * | myBamIndex |
| GenomeSequence * | myRefPtr |
| SamRecord::SequenceTranslation | myReadTranslation |
| SamRecord::SequenceTranslation | myWriteTranslation |
| std::string | myRefName |
Detailed Description
Allows the user to easily read/write a SAM/BAM file.
Definition at line 30 of file SamFile.h.
Member Enumeration Documentation
| enum SamFile::OpenType |
| enum SamFile::SortedType |
Enum for indicating the type of sort for the file.
- Enumerator:
UNSORTED file is not sorted.
FLAG SO flag from the header indicates the sort type.
COORDINATE file is sorted by coordinate.
QUERY_NAME file is sorted by queryname.
Definition at line 41 of file SamFile.h.
00041 { 00042 UNSORTED = 0, ///< file is not sorted. 00043 FLAG, ///< SO flag from the header indicates the sort type. 00044 COORDINATE, ///< file is sorted by coordinate. 00045 QUERY_NAME ///< file is sorted by queryname. 00046 };
Constructor & Destructor Documentation
| SamFile::SamFile | ( | ErrorHandler::HandlingType | errorHandlingType | ) |
Constructor that sets the error handling type.
- Parameters:
-
errorHandlingType how to handle errors.
Definition at line 42 of file SamFile.cpp.
References resetFile().
00043 : myFilePtr(NULL), 00044 myInterfacePtr(NULL), 00045 myStatistics(NULL), 00046 myStatus(errorHandlingType), 00047 myBamIndex(NULL), 00048 myRefPtr(NULL), 00049 myReadTranslation(SamRecord::NONE), 00050 myWriteTranslation(SamRecord::NONE) 00051 { 00052 resetFile(); 00053 }
| SamFile::SamFile | ( | const char * | filename, | |
| OpenType | mode | |||
| ) |
Constructor that opens the specified file based on the specified mode (READ/WRITE).
- Parameters:
-
filename name of the file to open. mode mode to use for opening the file.
Definition at line 58 of file SamFile.cpp.
00059 : myStatus() 00060 { 00061 init(filename, mode, NULL); 00062 }
| SamFile::SamFile | ( | const char * | filename, | |
| OpenType | mode, | |||
| ErrorHandler::HandlingType | errorHandlingType | |||
| ) |
Constructor that opens the specified file based on the specified mode (READ/WRITE) and handles errors per the specified handleType.
- Parameters:
-
filename name of the file to open. mode mode to use for opening the file. errorHandlingType how to handle errors.
Definition at line 67 of file SamFile.cpp.
00069 : myStatus(errorHandlingType) 00070 { 00071 init(filename, mode, NULL); 00072 }
| SamFile::SamFile | ( | const char * | filename, | |
| OpenType | mode, | |||
| SamFileHeader * | header | |||
| ) |
Constructor that opens the specified file based on the specified mode (READ/WRITE).
- Parameters:
-
filename name of the file to open. mode mode to use for opening the file. header to read into or write from
Definition at line 77 of file SamFile.cpp.
00078 : myStatus() 00079 { 00080 init(filename, mode, header); 00081 }
| SamFile::SamFile | ( | const char * | filename, | |
| OpenType | mode, | |||
| ErrorHandler::HandlingType | errorHandlingType, | |||
| SamFileHeader * | header | |||
| ) |
Constructor that opens the specified file based on the specified mode (READ/WRITE) and handles errors per the specified handleType.
- Parameters:
-
filename name of the file to open. mode mode to use for opening the file. errorHandlingType how to handle errors. header to read into or write from
Definition at line 86 of file SamFile.cpp.
00089 : myStatus(errorHandlingType) 00090 { 00091 init(filename, mode, header); 00092 }
Member Function Documentation
| void SamFile::GenerateStatistics | ( | bool | genStats | ) |
Whether or not statistics should be generated for this file.
The value is carried over between files and is not reset, but the statistics themselves are reset between files.
- Parameters:
-
genStats set to true if statistics should be generated, false if not.
Definition at line 832 of file SamFile.cpp.
References myStatistics.
00833 { 00834 if(genStats) 00835 { 00836 if(myStatistics == NULL) 00837 { 00838 // Want to generate statistics, but do not yet have the 00839 // structure for them, so create one. 00840 myStatistics = new SamStatistics(); 00841 } 00842 } 00843 else 00844 { 00845 // Do not generate statistics, so if myStatistics is not NULL, 00846 // delete it. 00847 if(myStatistics != NULL) 00848 { 00849 delete myStatistics; 00850 myStatistics = NULL; 00851 } 00852 } 00853 00854 }
| const BamIndex * SamFile::GetBamIndex | ( | ) |
Return the bam index if one has been opened.
- Returns:
- const pointer to the bam index, or null if one has not been opened.
Definition at line 857 of file SamFile.cpp.
| long int SamFile::GetCurrentPosition | ( | ) | [inline] |
| SamStatus::Status SamFile::GetFailure | ( | ) | [inline] |
Get the Status of the last call that sets status.
To remain backwards compatable - will be removed later.
Definition at line 175 of file SamFile.h.
References GetStatus().
00176 { 00177 return(GetStatus()); 00178 }
| int32_t SamFile::getNumMappedReadsFromIndex | ( | const char * | refName, | |
| SamFileHeader & | header | |||
| ) |
Get the number of mapped reads in the specified reference name.
Returns -1 for unknown reference names.
- Parameters:
-
refName reference name for which to extract the number of mapped reads. header header object containing the map from refName to refID
- Returns:
- number of mapped reads for the specified reference name.
Definition at line 774 of file SamFile.cpp.
References BamIndex::getNumMappedReads(), myStatus, and BamIndex::REF_ID_UNMAPPED.
00776 { 00777 // The bam index must have already been read. 00778 if(myBamIndex == NULL) 00779 { 00780 myStatus.setStatus(SamStatus::FAIL_ORDER, 00781 "Cannot get num mapped reads from the index until it has been read."); 00782 return(false); 00783 } 00784 int32_t refID = BamIndex::REF_ID_UNMAPPED; 00785 if((strcmp(refName, "") != 0) && (strcmp(refName, "*") != 0)) 00786 { 00787 // Reference name specified, so read just the "-1" entries. 00788 refID = header.getReferenceID(refName); 00789 } 00790 return(myBamIndex->getNumMappedReads(refID)); 00791 }
| int32_t SamFile::getNumMappedReadsFromIndex | ( | int32_t | refID | ) |
Get the number of mapped reads in the specified reference id.
Returns -1 for out of range refIDs.
- Parameters:
-
refID reference ID for which to extract the number of mapped reads.
- Returns:
- number of mapped reads for the specified reference id.
Definition at line 744 of file SamFile.cpp.
References BamIndex::getNumMappedReads(), and myStatus.
00745 { 00746 // The bam index must have already been read. 00747 if(myBamIndex == NULL) 00748 { 00749 myStatus.setStatus(SamStatus::FAIL_ORDER, 00750 "Cannot get num mapped reads from the index until it has been read."); 00751 return(false); 00752 } 00753 return(myBamIndex->getNumMappedReads(refID)); 00754 }
| uint32_t SamFile::GetNumOverlaps | ( | SamRecord & | samRecord | ) |
Returns the number of bases in the passed in read that overlap the region that is currently set.
- Parameters:
-
samRecord to check for overlapping bases.
- Returns:
- number of bases that overlap region that is currently set.
Definition at line 818 of file SamFile.cpp.
References SamRecord::getNumOverlaps(), SamRecord::setReference(), and SamRecord::setSequenceTranslation().
00819 { 00820 if(myRefPtr != NULL) 00821 { 00822 samRecord.setReference(myRefPtr); 00823 } 00824 samRecord.setSequenceTranslation(myReadTranslation); 00825 00826 // Get the overlaps in the sam record for the region currently set 00827 // for this file. 00828 return(samRecord.getNumOverlaps(myStartPos, myEndPos)); 00829 }
| int32_t SamFile::getNumUnMappedReadsFromIndex | ( | const char * | refName, | |
| SamFileHeader & | header | |||
| ) |
Get the number of unmapped reads in the specified reference name.
Returns -1 for unknown reference names.
- Parameters:
-
refName reference name for which to extract the number of unmapped reads. header header object containing the map from refName to refID
- Returns:
- number of unmapped reads for the specified reference name.
Definition at line 796 of file SamFile.cpp.
References BamIndex::getNumUnMappedReads(), myStatus, and BamIndex::REF_ID_UNMAPPED.
00798 { 00799 // The bam index must have already been read. 00800 if(myBamIndex == NULL) 00801 { 00802 myStatus.setStatus(SamStatus::FAIL_ORDER, 00803 "Cannot get num unmapped reads from the index until it has been read."); 00804 return(false); 00805 } 00806 int32_t refID = BamIndex::REF_ID_UNMAPPED; 00807 if((strcmp(refName, "") != 0) && (strcmp(refName, "*") != 0)) 00808 { 00809 // Reference name specified, so read just the "-1" entries. 00810 refID = header.getReferenceID(refName); 00811 } 00812 return(myBamIndex->getNumUnMappedReads(refID)); 00813 }
| int32_t SamFile::getNumUnMappedReadsFromIndex | ( | int32_t | refID | ) |
Get the number of unmapped reads in the specified reference id.
Returns -1 for out of range refIDs.
- Parameters:
-
refID reference ID for which to extract the number of unmapped reads.
- Returns:
- number of unmapped reads for the specified reference id.
Definition at line 759 of file SamFile.cpp.
References BamIndex::getNumUnMappedReads(), and myStatus.
00760 { 00761 // The bam index must have already been read. 00762 if(myBamIndex == NULL) 00763 { 00764 myStatus.setStatus(SamStatus::FAIL_ORDER, 00765 "Cannot get num unmapped reads from the index until it has been read."); 00766 return(false); 00767 } 00768 return(myBamIndex->getNumUnMappedReads(refID)); 00769 }
| bool SamFile::IsEOF | ( | ) |
Returns whether or not the end of the file has been reached.
- Returns:
- true = EOF; false = not eof. If the file is not open, false is returned.
Definition at line 425 of file SamFile.cpp.
References ifeof().
00426 { 00427 if (myFilePtr != NULL) 00428 { 00429 // File Pointer is set, so return if eof. 00430 return(ifeof(myFilePtr)); 00431 } 00432 // File pointer is not set, so return true, eof. 00433 return true; 00434 }
| bool SamFile::IsOpen | ( | ) |
Returns whether or not the file has been opened successfully.
- Returns:
- true = open; false = not open.
Definition at line 411 of file SamFile.cpp.
References InputFile::isOpen().
00412 { 00413 if (myFilePtr != NULL) 00414 { 00415 // File Pointer is set, so return if it is open. 00416 return(myFilePtr->isOpen()); 00417 } 00418 // File pointer is not set, so return false, not open. 00419 return false; 00420 }
| bool SamFile::OpenForRead | ( | const char * | filename, | |
| SamFileHeader * | header = NULL | |||
| ) |
Open a sam/bam file for reading with the specified filename.
- Parameters:
-
filename the sam/bam file to open for reading. header to read into or write from (optional)
- Returns:
- true = success; false = failure.
Definition at line 106 of file SamFile.cpp.
References InputFile::BGZF, InputFile::DEFAULT, ifopen(), ifread(), ifrewind(), myIsBamOpenForRead, myIsOpenForRead, myStatus, ReadHeader(), resetFile(), InputFile::setAttemptRecovery(), and InputFile::UNCOMPRESSED.
00107 { 00108 // Reset for any previously operated on files. 00109 resetFile(); 00110 00111 int lastchar = 0; 00112 00113 while (filename[lastchar] != 0) lastchar++; 00114 00115 // If at least one character, check for '-'. 00116 if((lastchar >= 1) && (filename[0] == '-')) 00117 { 00118 // Read from stdin - determine type of file to read. 00119 // Determine if compressed bam. 00120 if(strcmp(filename, "-.bam") == 0) 00121 { 00122 // Compressed bam - open as bgzf. 00123 // -.bam is the filename, read compressed bam from stdin 00124 filename = "-"; 00125 00126 myFilePtr = new InputFile; 00127 // support recover mode - this switches in a reader 00128 // capable of recovering from bad BGZF compression blocks. 00129 myFilePtr->setAttemptRecovery(myAttemptRecovery); 00130 myFilePtr->openFile(filename, "rb", InputFile::BGZF); 00131 00132 myInterfacePtr = new BamInterface; 00133 00134 // Read the magic string. 00135 char magic[4]; 00136 ifread(myFilePtr, magic, 4); 00137 } 00138 else if(strcmp(filename, "-.ubam") == 0) 00139 { 00140 // uncompressed BAM File. 00141 // -.ubam is the filename, read uncompressed bam from stdin. 00142 // uncompressed BAM is still compressed with BGZF, but using 00143 // compression level 0, so still open as BGZF since it has a 00144 // BGZF header. 00145 filename = "-"; 00146 00147 // Uncompressed, so do not require the eof block. 00148 BgzfFileType::setRequireEofBlock(false); 00149 00150 myFilePtr = ifopen(filename, "rb", InputFile::BGZF); 00151 00152 myInterfacePtr = new BamInterface; 00153 00154 // Read the magic string. 00155 char magic[4]; 00156 ifread(myFilePtr, magic, 4); 00157 } 00158 else 00159 { 00160 // SAM File. 00161 // read sam from stdin 00162 filename = "-"; 00163 myFilePtr = ifopen(filename, "rb", InputFile::UNCOMPRESSED); 00164 myInterfacePtr = new SamInterface; 00165 } 00166 } 00167 else 00168 { 00169 // Not from stdin. Read the file to determine the type. 00170 00171 myFilePtr = new InputFile; 00172 00173 // support recovery mode - this conditionally enables a reader 00174 // capable of recovering from bad BGZF compression blocks. 00175 myFilePtr->setAttemptRecovery(myAttemptRecovery); 00176 bool rc = myFilePtr->openFile(filename, "rb", InputFile::DEFAULT); 00177 00178 if (rc == false) 00179 { 00180 std::string errorMessage = "Failed to Open "; 00181 errorMessage += filename; 00182 errorMessage += " for reading"; 00183 myStatus.setStatus(SamStatus::FAIL_IO, errorMessage.c_str()); 00184 delete myFilePtr; 00185 return(false); 00186 } 00187 00188 char magic[4]; 00189 ifread(myFilePtr, magic, 4); 00190 00191 if (magic[0] == 'B' && magic[1] == 'A' && magic[2] == 'M' && 00192 magic[3] == 1) 00193 { 00194 myInterfacePtr = new BamInterface; 00195 // Set that it is a bam file open for reading. This is needed to 00196 // determine if an index file can be used. 00197 myIsBamOpenForRead = true; 00198 } 00199 else 00200 { 00201 // Not a bam, so rewind to the beginning of the file so it 00202 // can be read. 00203 ifrewind(myFilePtr); 00204 myInterfacePtr = new SamInterface; 00205 } 00206 } 00207 00208 // File is open for reading. 00209 myIsOpenForRead = true; 00210 00211 // Read the header if one was passed in. 00212 if(header != NULL) 00213 { 00214 return(ReadHeader(*header)); 00215 } 00216 00217 // Successfully opened the file. 00218 myStatus = SamStatus::SUCCESS; 00219 return(true); 00220 }
| bool SamFile::OpenForWrite | ( | const char * | filename, | |
| SamFileHeader * | header = NULL | |||
| ) |
Open a sam/bam file for writing with the specified filename.
- Parameters:
-
filename the sam/bam file to open for writing. header to read into or write from (optional)
- Returns:
- true = success; false = failure.
Definition at line 224 of file SamFile.cpp.
References InputFile::BGZF, ifopen(), myIsOpenForWrite, myStatus, resetFile(), InputFile::UNCOMPRESSED, and WriteHeader().
00225 { 00226 // Reset for any previously operated on files. 00227 resetFile(); 00228 00229 int lastchar = 0; 00230 while (filename[lastchar] != 0) lastchar++; 00231 if (lastchar >= 4 && 00232 filename[lastchar - 4] == 'u' && 00233 filename[lastchar - 3] == 'b' && 00234 filename[lastchar - 2] == 'a' && 00235 filename[lastchar - 1] == 'm') 00236 { 00237 // BAM File. 00238 // if -.ubam is the filename, write uncompressed bam to stdout 00239 if((lastchar == 6) && (filename[0] == '-') && (filename[1] == '.')) 00240 { 00241 filename = "-"; 00242 } 00243 00244 myFilePtr = ifopen(filename, "wb0", InputFile::BGZF); 00245 00246 myInterfacePtr = new BamInterface; 00247 } 00248 else if (lastchar >= 3 && 00249 filename[lastchar - 3] == 'b' && 00250 filename[lastchar - 2] == 'a' && 00251 filename[lastchar - 1] == 'm') 00252 { 00253 // BAM File. 00254 // if -.bam is the filename, write compressed bam to stdout 00255 if((lastchar == 5) && (filename[0] == '-') && (filename[1] == '.')) 00256 { 00257 filename = "-"; 00258 } 00259 myFilePtr = ifopen(filename, "wb", InputFile::BGZF); 00260 00261 myInterfacePtr = new BamInterface; 00262 } 00263 else 00264 { 00265 // SAM File 00266 // if - (followed by anything is the filename, 00267 // write uncompressed sam to stdout 00268 if((lastchar >= 1) && (filename[0] == '-')) 00269 { 00270 filename = "-"; 00271 } 00272 myFilePtr = ifopen(filename, "wb", InputFile::UNCOMPRESSED); 00273 00274 myInterfacePtr = new SamInterface; 00275 } 00276 00277 if (myFilePtr == NULL) 00278 { 00279 std::string errorMessage = "Failed to Open "; 00280 errorMessage += filename; 00281 errorMessage += " for writing"; 00282 myStatus.setStatus(SamStatus::FAIL_IO, errorMessage.c_str()); 00283 return(false); 00284 } 00285 00286 myIsOpenForWrite = true; 00287 00288 // Write the header if one was passed in. 00289 if(header != NULL) 00290 { 00291 return(WriteHeader(*header)); 00292 } 00293 00294 // Successfully opened the file. 00295 myStatus = SamStatus::SUCCESS; 00296 return(true); 00297 }
| bool SamFile::ReadBamIndex | ( | ) |
Read the bam index file using the BAM filename as a base.
It must be read prior to setting a read section, for seeking and reading portions of a bam file. Must be read after opening the BAM file since it uses the BAM filename as a base name for the index file. First it tries filename.bam.bai. If that fails, it tries it without the .bam extension, filename.bai.
- Returns:
- true = success; false = failure.
Definition at line 329 of file SamFile.cpp.
References InputFile::getFileName(), and myStatus.
00330 { 00331 if(myFilePtr == NULL) 00332 { 00333 // Can't read the bam index file because the BAM file has not yet been 00334 // opened, so we don't know the base filename for the index file. 00335 std::string errorMessage = "Failed to read the bam Index file -" 00336 " the BAM file needs to be read first in order to determine" 00337 " the index filename."; 00338 myStatus.setStatus(SamStatus::FAIL_ORDER, errorMessage.c_str()); 00339 return(false); 00340 } 00341 00342 const char* bamBaseName = myFilePtr->getFileName(); 00343 00344 std::string indexName = bamBaseName; 00345 indexName += ".bai"; 00346 00347 bool foundFile = true; 00348 try 00349 { 00350 if(ReadBamIndex(indexName.c_str()) == false) 00351 { 00352 foundFile = false; 00353 } 00354 } 00355 catch (std::exception& e) 00356 { 00357 foundFile = false; 00358 } 00359 00360 // Check to see if the index file was found. 00361 if(!foundFile) 00362 { 00363 // Not found - try without the bam extension. 00364 // Locate the start of the bam extension 00365 size_t startExt = indexName.find(".bam"); 00366 if(startExt == std::string::npos) 00367 { 00368 // Could not find the .bam extension, so just return false since the 00369 // call to ReadBamIndex set the status. 00370 return(false); 00371 } 00372 // Remove ".bam" and try reading the index again. 00373 indexName.erase(startExt, 4); 00374 return(ReadBamIndex(indexName.c_str())); 00375 } 00376 return(true); 00377 }
| bool SamFile::ReadBamIndex | ( | const char * | filename | ) |
Read the specified bam index file.
It must be read prior to setting a read section, for seeking and reading portions of a bam file.
- Parameters:
-
filename the name of the bam index file to be read.
- Returns:
- true = success; false = failure.
Definition at line 301 of file SamFile.cpp.
References myStatus, and BamIndex::readIndex().
00302 { 00303 // Cleanup a previously setup index. 00304 if(myBamIndex != NULL) 00305 { 00306 delete myBamIndex; 00307 myBamIndex = NULL; 00308 } 00309 00310 // Create a new bam index. 00311 myBamIndex = new BamIndex(); 00312 SamStatus::Status indexStat = myBamIndex->readIndex(bamIndexFilename); 00313 00314 if(indexStat != SamStatus::SUCCESS) 00315 { 00316 std::string errorMessage = "Failed to read the bam Index file: "; 00317 errorMessage += bamIndexFilename; 00318 myStatus.setStatus(indexStat, errorMessage.c_str()); 00319 delete myBamIndex; 00320 myBamIndex = NULL; 00321 return(false); 00322 } 00323 myStatus = SamStatus::SUCCESS; 00324 return(true); 00325 }
| bool SamFile::ReadHeader | ( | SamFileHeader & | header | ) |
Reads the header section from the file and stores it in the passed in header.
- Returns:
- true = success; false = failure.
Definition at line 438 of file SamFile.cpp.
References myHasHeader, myIsOpenForRead, and myStatus.
Referenced by OpenForRead().
00439 { 00440 if(myIsOpenForRead == false) 00441 { 00442 // File is not open for read 00443 myStatus.setStatus(SamStatus::FAIL_ORDER, 00444 "Cannot read header since the file is not open for reading"); 00445 return(false); 00446 } 00447 00448 if(myHasHeader == true) 00449 { 00450 // The header has already been read. 00451 myStatus.setStatus(SamStatus::FAIL_ORDER, 00452 "Cannot read header since it has already been read."); 00453 return(false); 00454 } 00455 00456 myStatus = myInterfacePtr->readHeader(myFilePtr, header); 00457 if(myStatus == SamStatus::SUCCESS) 00458 { 00459 // The header has now been successfully read. 00460 myHasHeader = true; 00461 return(true); 00462 } 00463 return(false); 00464 }
| bool SamFile::ReadRecord | ( | SamFileHeader & | header, | |
| SamRecord & | record | |||
| ) |
Reads the next record from the file & stores it in the passed in record.
- Returns:
- true = record was successfully set. false = record was not successfully set.
Definition at line 502 of file SamFile.cpp.
References myHasHeader, myIsOpenForRead, myRecordCount, myStatistics, myStatus, readIndexedRecord(), BamIndex::REF_ID_ALL, SamRecord::setReference(), SamRecord::setSequenceTranslation(), and validateSortOrder().
00504 { 00505 myStatus = SamStatus::SUCCESS; 00506 00507 if(myIsOpenForRead == false) 00508 { 00509 // File is not open for read 00510 myStatus.setStatus(SamStatus::FAIL_ORDER, 00511 "Cannot read record since the file is not open for reading"); 00512 throw(std::runtime_error("SOFTWARE BUG: trying to read a SAM/BAM record prior to opening the file.")); 00513 return(false); 00514 } 00515 00516 if(myHasHeader == false) 00517 { 00518 // The header has not yet been read. 00519 // TODO - maybe just read the header. 00520 myStatus.setStatus(SamStatus::FAIL_ORDER, 00521 "Cannot read record since the header has not been read."); 00522 throw(std::runtime_error("SOFTWARE BUG: trying to read a SAM/BAM record prior to reading the header.")); 00523 return(false); 00524 } 00525 00526 // Check to see if a new region has been set. If so, determine the 00527 // chunks for that region. 00528 if(myNewSection) 00529 { 00530 if(!processNewSection(header)) 00531 { 00532 // Failed processing a new section. Could be an 00533 // order issue like the file not being open or the 00534 // indexed file not having been read. 00535 // processNewSection sets myStatus with the failure reason. 00536 return(false); 00537 } 00538 } 00539 00540 // Check to see if the file should be read by index. 00541 if(myRefID != BamIndex::REF_ID_ALL) 00542 { 00543 // Reference ID is set, so read by index. 00544 return(readIndexedRecord(header, record)); 00545 } 00546 00547 record.setReference(myRefPtr); 00548 record.setSequenceTranslation(myReadTranslation); 00549 00550 // File is open for reading and the header has been read, so read the next 00551 // record. 00552 myInterfacePtr->readRecord(myFilePtr, header, record, myStatus); 00553 if(myStatus == SamStatus::SUCCESS) 00554 { 00555 // A record was successfully read, so increment the record count. 00556 myRecordCount++; 00557 00558 if(myStatistics != NULL) 00559 { 00560 // Statistics should be updated. 00561 myStatistics->updateStatistics(record); 00562 } 00563 00564 // Successfully read the record, so check the sort order. 00565 if(!validateSortOrder(record, header)) 00566 { 00567 // ValidateSortOrder sets the status on a failure. 00568 return(false); 00569 } 00570 return(true); 00571 } 00572 // Failed to read the record. 00573 return(false); 00574 }
| bool SamFile::SetReadSection | ( | const char * | refName, | |
| int32_t | start, | |||
| int32_t | end, | |||
| bool | overlap = true | |||
| ) |
Sets what part of the BAM file should be read.
This version will set it to only read a specific reference name and start/end position. The records for this section will be retrieved on each ReadRecord call. When all records have been retrieved for the specified section, ReadRecord will return failure until a new read section is set. Must be called only after the file has been opened for reading. Sorting validation is reset everytime SetReadPosition is called since it can jump around in the file.
- Parameters:
-
refName the reference name of the records to read from the file. start inclusive 0-based start position of records that should be read for this refID. end exclusive 0-based end position of records that should be read for this refID. overlap When true (default), return reads that just overlap the region. When false, only return reads that fall completely within the region
- Returns:
- true = success; false = failure.
Definition at line 696 of file SamFile.cpp.
References myIsBamOpenForRead, myPrevCoord, myStatus, BamIndex::REF_ID_ALL, and BamIndex::REF_ID_UNMAPPED.
00698 { 00699 // If there is not a BAM file open for reading, return failure. 00700 // Opening a new file clears the read section, so it must be 00701 // set after the file is opened. 00702 if(!myIsBamOpenForRead) 00703 { 00704 // There is not a BAM file open for reading. 00705 myStatus.setStatus(SamStatus::FAIL_ORDER, 00706 "Cannot set section since there is no bam file open"); 00707 return(false); 00708 } 00709 00710 myNewSection = true; 00711 myOverlapSection = overlap; 00712 myStartPos = start; 00713 myEndPos = end; 00714 if((strcmp(refName, "") == 0) || (strcmp(refName, "*") == 0)) 00715 { 00716 // No Reference name specified, so read just the "-1" entries. 00717 myRefID = BamIndex::REF_ID_UNMAPPED; 00718 } 00719 else 00720 { 00721 // save the reference name and revert the reference ID to unknown 00722 // so it will be calculated later. 00723 myRefName = refName; 00724 myRefID = BamIndex::REF_ID_ALL; 00725 } 00726 myChunksToRead.clear(); 00727 // Reset the end of the current chunk. We are resetting our read, so 00728 // we no longer have a "current chunk" that we are reading. 00729 myCurrentChunkEnd = 0; 00730 myStatus = SamStatus::SUCCESS; 00731 00732 // Reset the sort order criteria since we moved around in the file. 00733 myPrevCoord = -1; 00734 myPrevRefID = 0; 00735 myPrevReadName.clear(); 00736 00737 return(true); 00738 }
| bool SamFile::SetReadSection | ( | int32_t | refID, | |
| int32_t | start, | |||
| int32_t | end, | |||
| bool | overlap = true | |||
| ) |
Sets what part of the BAM file should be read.
This version will set it to only read a specific reference id and start/end position. The records for this section will be retrieved on each ReadRecord call. When all records have been retrieved for the specified section, ReadRecord will return failure until a new read section is set. Must be called only after the file has been opened for reading. Sorting validation is reset everytime SetReadPosition is called since it can jump around in the file.
- Parameters:
-
refID the reference ID of the records to read from the file. start inclusive 0-based start position of records that should be read for this refID. end exclusive 0-based end position of records that should be read for this refID. overlap When true (default), return reads that just overlap the region. When false, only return reads that fall completely within the region
- Returns:
- true = success; false = failure.
Definition at line 660 of file SamFile.cpp.
References myIsBamOpenForRead, myPrevCoord, and myStatus.
00662 { 00663 // If there is not a BAM file open for reading, return failure. 00664 // Opening a new file clears the read section, so it must be 00665 // set after the file is opened. 00666 if(!myIsBamOpenForRead) 00667 { 00668 // There is not a BAM file open for reading. 00669 myStatus.setStatus(SamStatus::FAIL_ORDER, 00670 "Cannot set section since there is no bam file open"); 00671 return(false); 00672 } 00673 00674 myNewSection = true; 00675 myOverlapSection = overlap; 00676 myStartPos = start; 00677 myEndPos = end; 00678 myRefID = refID; 00679 myRefName.clear(); 00680 myChunksToRead.clear(); 00681 // Reset the end of the current chunk. We are resetting our read, so 00682 // we no longer have a "current chunk" that we are reading. 00683 myCurrentChunkEnd = 0; 00684 myStatus = SamStatus::SUCCESS; 00685 00686 // Reset the sort order criteria since we moved around in the file. 00687 myPrevCoord = -1; 00688 myPrevRefID = 0; 00689 myPrevReadName.clear(); 00690 00691 return(true); 00692 }
| bool SamFile::SetReadSection | ( | const char * | refName | ) |
Sets what part of the BAM file should be read.
This version will set it to only read a specific reference name. The records for that reference id will be retrieved on each ReadRecord call. When all records have been retrieved for the specified reference name, ReadRecord will return failure until a new read section is set. Must be called only after the file has been opened for reading. Sorting validation is reset everytime SetReadPosition is called since it can jump around in the file.
- Parameters:
-
refName the reference name of the records to read from the file.
- Returns:
- true = success; false = failure.
Definition at line 652 of file SamFile.cpp.
References SetReadSection().
00653 { 00654 // No start/end specified, so set back to default -1. 00655 return(SetReadSection(refName, -1, -1)); 00656 }
| bool SamFile::SetReadSection | ( | int32_t | refID | ) |
Sets what part of the BAM file should be read.
This version will set it to only read a specific reference id. The records for that reference id will be retrieved on each ReadRecord call. When all records have been retrieved for the specified reference id, ReadRecord will return failure until a new read section is set. Must be called only after the file has been opened for reading. Sorting validation is reset everytime SetReadPosition is called since it can jump around in the file.
- Parameters:
-
refID the reference ID of the records to read from the file.
- Returns:
- true = success; false = failure.
Definition at line 643 of file SamFile.cpp.
Referenced by SetReadSection().
00644 { 00645 // No start/end specified, so set back to default -1. 00646 return(SetReadSection(refID, -1, -1)); 00647 }
| void SamFile::SetReadSequenceTranslation | ( | SamRecord::SequenceTranslation | translation | ) |
Set the type of sequence translation to use when reading the sequence.
Passed down to the SamRecord when it is read. NONE (the sequence is left as-is).
- Parameters:
-
translation type of sequence translation to use.
Definition at line 388 of file SamFile.cpp.
| void SamFile::SetReference | ( | GenomeSequence * | reference | ) |
Sets the reference to the specified genome sequence object.
- Parameters:
-
reference pointer to the GenomeSequence object.
Definition at line 381 of file SamFile.cpp.
| void SamFile::setSortedValidation | ( | SortedType | sortType | ) |
Set the flag to validate that the file is sorted as it is read/written.
Must be called after the file has been opened. Sorting validation is reset everytime SetReadPosition is called since it can jump around in the file.
Definition at line 629 of file SamFile.cpp.
| void SamFile::SetWriteSequenceTranslation | ( | SamRecord::SequenceTranslation | translation | ) |
Set the type of sequence translation to use when writing the sequence.
Passed down to the SamRecord when it is written. The default type (if this method is never called) is NONE (the sequence is left as-is).
- Parameters:
-
translation type of sequence translation to use.
Definition at line 395 of file SamFile.cpp.
| bool SamFile::validateSortOrder | ( | SamRecord & | record, | |
| SamFileHeader & | header | |||
| ) | [protected] |
Validate that the record is sorted compared to the previously read record if there is one, according to the specified sort order.
If the sort order is UNSORTED, true is returned. Sorting validation is reset everytime SetReadPosition is called since it can jump around in the file.
Definition at line 951 of file SamFile.cpp.
References FLAG, SamRecord::get0BasedPosition(), SamRecord::getReadName(), SamRecord::getReferenceID(), myPrevCoord, myRecordCount, myStatus, QUERY_NAME, BamIndex::REF_ID_UNMAPPED, SamRecord::setReference(), SamRecord::setSequenceTranslation(), and UNSORTED.
Referenced by readIndexedRecord(), ReadRecord(), and WriteRecord().
00952 { 00953 if(myRefPtr != NULL) 00954 { 00955 record.setReference(myRefPtr); 00956 } 00957 record.setSequenceTranslation(myReadTranslation); 00958 00959 bool status = false; 00960 if(mySortedType == UNSORTED) 00961 { 00962 // Unsorted, so nothing to validate, just return true. 00963 status = true; 00964 } 00965 else 00966 { 00967 // Check to see if mySortedType is based on the header. 00968 if(mySortedType == FLAG) 00969 { 00970 // Determine the sorted type from what was read out of the header. 00971 mySortedType = getSortOrderFromHeader(header); 00972 } 00973 00974 if(mySortedType == QUERY_NAME) 00975 { 00976 // Validate that it is sorted by query name. 00977 // Get the query name from the record. 00978 const char* readName = record.getReadName(); 00979 if(myPrevReadName.compare(readName) > 0) 00980 { 00981 // The previous name is greater than the new record's name, so 00982 // return false. 00983 String errorMessage = "ERROR: File is not sorted at record "; 00984 errorMessage += myRecordCount; 00985 myStatus.setStatus(SamStatus::INVALID_SORT, 00986 errorMessage.c_str()); 00987 status = false; 00988 } 00989 else 00990 { 00991 myPrevReadName = readName; 00992 status = true; 00993 } 00994 } 00995 else 00996 { 00997 // Validate that it is sorted by COORDINATES. 00998 // Get the leftmost coordinate and the reference index. 00999 int32_t refID = record.getReferenceID(); 01000 int32_t coord = record.get0BasedPosition(); 01001 // The unmapped reference id is at the end of a sorted file. 01002 if(refID == BamIndex::REF_ID_UNMAPPED) 01003 { 01004 // A new reference ID that is for the unmapped reads 01005 // is always valid. 01006 status = true; 01007 myPrevRefID = refID; 01008 myPrevCoord = coord; 01009 } 01010 else if(myPrevRefID == BamIndex::REF_ID_UNMAPPED) 01011 { 01012 // Previous reference ID was for unmapped reads, but the 01013 // current one is not, so this is not sorted. 01014 String errorMessage = "ERROR: File is not sorted at record "; 01015 errorMessage += myRecordCount; 01016 myStatus.setStatus(SamStatus::INVALID_SORT, 01017 errorMessage.c_str()); 01018 status = false; 01019 } 01020 else if(refID < myPrevRefID) 01021 { 01022 // Current reference id is less than the previous one, 01023 //meaning that it is not sorted. 01024 String errorMessage = "ERROR: File is not sorted at record "; 01025 errorMessage += myRecordCount; 01026 myStatus.setStatus(SamStatus::INVALID_SORT, 01027 errorMessage.c_str()); 01028 status = false; 01029 } 01030 else 01031 { 01032 // The reference IDs are in the correct order. 01033 if(refID > myPrevRefID) 01034 { 01035 // New reference id, so set the previous coordinate to -1 01036 myPrevCoord = -1; 01037 } 01038 01039 // Check the coordinates. 01040 if(coord < myPrevCoord) 01041 { 01042 // New Coord is less than the previous position. 01043 String errorMessage = "ERROR: File is not sorted at record "; 01044 errorMessage += myRecordCount; 01045 myStatus.setStatus(SamStatus::INVALID_SORT, 01046 errorMessage.c_str()); 01047 status = false; 01048 } 01049 else 01050 { 01051 myPrevRefID = refID; 01052 myPrevCoord = coord; 01053 status = true; 01054 } 01055 } 01056 } 01057 } 01058 01059 return(status); 01060 }
| bool SamFile::WriteHeader | ( | SamFileHeader & | header | ) |
Writes the specified header into the file.
- Returns:
- true = success; false = failure.
Definition at line 468 of file SamFile.cpp.
References myHasHeader, myIsOpenForWrite, and myStatus.
Referenced by OpenForWrite().
00469 { 00470 if(myIsOpenForWrite == false) 00471 { 00472 // File is not open for write 00473 // -OR- 00474 // The header has already been written. 00475 myStatus.setStatus(SamStatus::FAIL_ORDER, 00476 "Cannot write header since the file is not open for writing"); 00477 return(false); 00478 } 00479 00480 if(myHasHeader == true) 00481 { 00482 // The header has already been written. 00483 myStatus.setStatus(SamStatus::FAIL_ORDER, 00484 "Cannot write header since it has already been written"); 00485 return(false); 00486 } 00487 00488 myStatus = myInterfacePtr->writeHeader(myFilePtr, header); 00489 if(myStatus == SamStatus::SUCCESS) 00490 { 00491 // The header has now been successfully written. 00492 myHasHeader = true; 00493 return(true); 00494 } 00495 00496 // return the status. 00497 return(false); 00498 }
| bool SamFile::WriteRecord | ( | SamFileHeader & | header, | |
| SamRecord & | record | |||
| ) |
Writes the specified record into the file.
- Returns:
- true = success; false = failure.
Definition at line 579 of file SamFile.cpp.
References myHasHeader, myIsOpenForWrite, myRecordCount, myStatus, SamRecord::setReference(), and validateSortOrder().
00581 { 00582 if(myIsOpenForWrite == false) 00583 { 00584 // File is not open for writing 00585 myStatus.setStatus(SamStatus::FAIL_ORDER, 00586 "Cannot write record since the file is not open for writing"); 00587 return(false); 00588 } 00589 00590 if(myHasHeader == false) 00591 { 00592 // The header has not yet been written. 00593 myStatus.setStatus(SamStatus::FAIL_ORDER, 00594 "Cannot write record since the header has not been written"); 00595 return(false); 00596 } 00597 00598 // Before trying to write the record, validate the sort order. 00599 if(!validateSortOrder(record, header)) 00600 { 00601 // Not sorted like it is supposed to be, do not write the record 00602 myStatus.setStatus(SamStatus::INVALID_SORT, 00603 "Cannot write the record since the file is not properly sorted."); 00604 return(false); 00605 } 00606 00607 if(myRefPtr != NULL) 00608 { 00609 record.setReference(myRefPtr); 00610 } 00611 00612 // File is open for writing and the header has been written, so write the 00613 // record. 00614 myStatus = myInterfacePtr->writeRecord(myFilePtr, header, record, 00615 myWriteTranslation); 00616 00617 if(myStatus == SamStatus::SUCCESS) 00618 { 00619 // A record was successfully written, so increment the record count. 00620 myRecordCount++; 00621 return(true); 00622 } 00623 return(false); 00624 }
Member Data Documentation
bool SamFile::myHasHeader [protected] |
Flag to indicate if a header has been read/written - required before being able to read/write a record.
Definition at line 341 of file SamFile.h.
Referenced by ReadHeader(), ReadRecord(), resetFile(), WriteHeader(), and WriteRecord().
The documentation for this class was generated from the following files:
- bam/SamFile.h
- bam/SamFile.cpp
 1.6.3
1.6.3