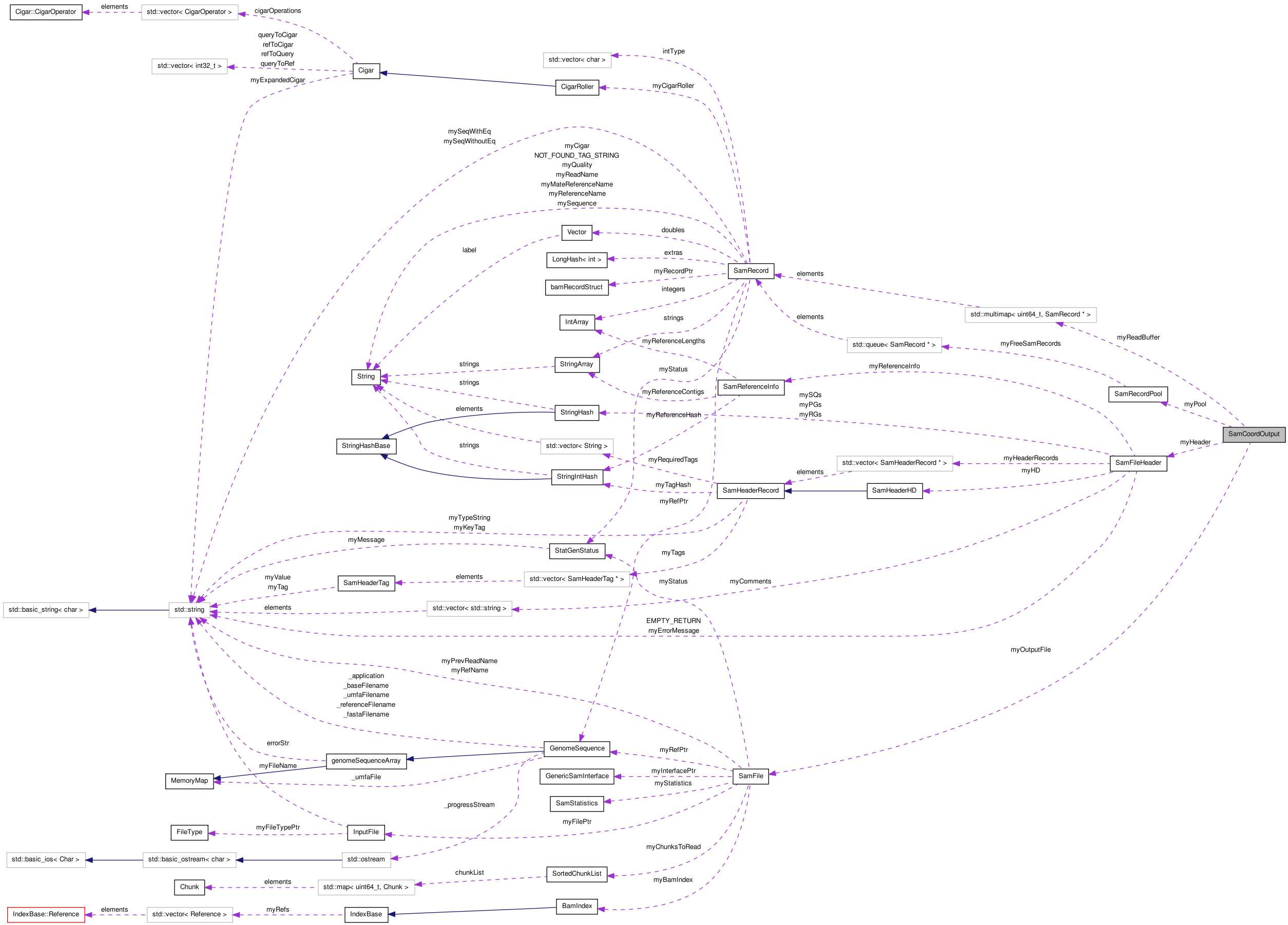
SamCoordOutput Class Reference
Class for buffering up output reads to ensure that it is sorted. More...
#include <SamCoordOutput.h>
Public Member Functions | |
| SamCoordOutput (SamRecordPool &pool) | |
| Create an output buffer returning any written records to the specified pool. | |
| void | setOutputFile (SamFile *outFile, SamFileHeader *header) |
| Set the already opened output file to write to when flushed. | |
| bool | add (SamRecord *record) |
| Add the specified record to this read buffer. | |
| bool | flushAll () |
| Flush the entire buffer, writing all records. | |
| bool | flush (int32_t chromID, int32_t pos0Based) |
| Flush the buffer based on the specified chromosome id/position, writing any records that start at/before the specified chromosome id/position. | |
Detailed Description
Class for buffering up output reads to ensure that it is sorted.
They are added in almost sorted order. Flush writes any records that start at/before the specified position.
Definition at line 27 of file SamCoordOutput.h.
Constructor & Destructor Documentation
| SamCoordOutput::SamCoordOutput | ( | SamRecordPool & | pool | ) |
Create an output buffer returning any written records to the specified pool.
- Parameters:
-
pool pool that any written records should be returned to, a pointer to this pool is stored, so it should not go out of scope until the output buffer has written all the records.
Definition at line 22 of file SamCoordOutput.cpp.
References SamCoordOutput().
Referenced by SamCoordOutput().
Member Function Documentation
| bool SamCoordOutput::flush | ( | int32_t | chromID, | |
| int32_t | pos0Based | |||
| ) |
Flush the buffer based on the specified chromosome id/position, writing any records that start at/before the specified chromosome id/position.
If no output buffer is set, the files cannot be written, but the flushed records are removed/freed. A chromID of -1 will flush everything regardless of pos0Based.
Definition at line 64 of file SamCoordOutput.cpp.
References SamHelper::combineChromPos(), SamRecordPool::releaseRecord(), and SamFile::WriteRecord().
Referenced by flushAll().
00065 { 00066 static std::multimap<uint64_t, SamRecord*>::iterator iter; 00067 00068 uint64_t chromPos = SamHelper::combineChromPos(chromID, pos0Based); 00069 00070 bool returnVal = true; 00071 iter = myReadBuffer.begin(); 00072 00073 if((myOutputFile == NULL) || (myHeader == NULL)) 00074 { 00075 std::cerr << 00076 "SamCoordOutput::flush, no output file/header is set, so records removed without being written\n"; 00077 returnVal = false; 00078 } 00079 00080 while((iter != myReadBuffer.end()) && 00081 (((*iter).first <= chromPos) || (chromID == -1))) 00082 { 00083 if((myOutputFile != NULL) && (myHeader != NULL)) 00084 { 00085 returnVal &= 00086 myOutputFile->WriteRecord(*myHeader, *((*iter).second)); 00087 } 00088 if(myPool != NULL) 00089 { 00090 myPool->releaseRecord((*iter).second); 00091 } 00092 else 00093 { 00094 delete((*iter).second); 00095 } 00096 ++iter; 00097 } 00098 // Remove the elements from the begining up to, 00099 // but not including the current iterator position. 00100 myReadBuffer.erase(myReadBuffer.begin(), iter); 00101 00102 return(returnVal); 00103 }
| bool SamCoordOutput::flushAll | ( | ) |
Flush the entire buffer, writing all records.
If no output buffer is set, the files cannot be written, but the flushed records are removed/freed.
Definition at line 59 of file SamCoordOutput.cpp.
References flush().
00060 { 00061 return(flush(-1,-1)); 00062 }
| void SamCoordOutput::setOutputFile | ( | SamFile * | outFile, | |
| SamFileHeader * | header | |||
| ) |
Set the already opened output file to write to when flushed.
The user should not close/delete the SamFile until this class is done with it. This class does NOT close/delete the SamFile.
- Parameters:
-
outFile pointer to an already opened (and header written) SAM/BAM output file. header pointer to an already written header that should be used for writing the records.
Definition at line 38 of file SamCoordOutput.cpp.
The documentation for this class was generated from the following files:
- bam/SamCoordOutput.h
- bam/SamCoordOutput.cpp
 1.6.3
1.6.3