When the --hardyWeinberg option is specified, PEDSTATS will check all markers in your
data file to see if genotype frequencies within your sample appear to deviate significantly
from Hardy-Weinberg equilibrium. In this section, we'll discuss the test statistics implemented
in PEDSTATS and look at some text-based output produced by the program when Hardy-Weinberg checks
are run. If you're already familiar with Hardy-Weinberg testing and have used this option in previous
versions of PEDSTATS, you may want still want to take a look at the sections on our fast exact test for SNPs, Hardy-Weinberg selection strategies and graphical
summaries of Hardy-Weinberg tests.
- Allele frequencies (pi) are determined by allele counting
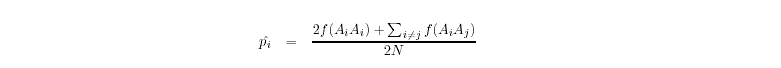
- All alleles are classified as either rare or common:
| Rare | Common |
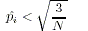
|
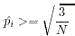
|
- If all alleles are rare, the rare allele with highest frequency is removed from the pool of rare of
alleles and placed in a pool on its own (this ensures no tests are attempted with only one pool)
- All alleles in the rare pool
 are grouped
into a single cell and the grouped allele frequency
are grouped
into a single cell and the grouped allele frequency 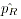 is again calculated via allele counting.
is again calculated via allele counting.
- If
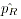 is still less than
is still less than 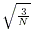 and there is more than one common allele,
PEDSTATS augments the pool of rare alleles R by
adding the common allele with minimum frequency.
and there is more than one common allele,
PEDSTATS augments the pool of rare alleles R by
adding the common allele with minimum frequency.
Once the pooling algorithm runs, PEDSTATS will perform a chi-squared test on any markers
with more than two allele pools. Any markers with two allele pools will be tested using
an exact test of Hardy-Weinberg equilibrium.
Asymptotic chi-squared test
For the chi-squared test, PEDSTATS uses the test statistic
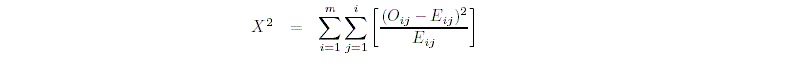 where m denotes the number of alleles at the marker locus, Ai
denotes the ith allele, Eij denotes the expected
frequency of the genotype AiAj and Oij denotes
the observed frequency of AiAj.
where m denotes the number of alleles at the marker locus, Ai
denotes the ith allele, Eij denotes the expected
frequency of the genotype AiAj and Oij denotes
the observed frequency of AiAj.
Under the null hypothesis of Hardy-Weinberg equilibrium,
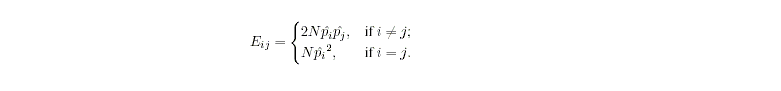 where N is the number of
individuals in your sample that have been genotyped at the locus and pi
is the allele frequency estimate for Ai obtained via allele counting.
where N is the number of
individuals in your sample that have been genotyped at the locus and pi
is the allele frequency estimate for Ai obtained via allele counting.
When expected counts are large, X2 will have an approximate chi-squared
distribution with m(m-1)/2 degrees of freedom for a marker with m alleles. When
fitted values are unreasonably small (e.g, Eij < 3), X2
will not have an approximate chi-squared distribution and can be somewhat unreliable. In this case,
PEDSTATS will still perform the test but the result will be flagged.
Text Output for Hardy-Weinberg Tests
If you want to take a look at some output for this option, in the examples subdirectory,
try running:
pedstats -p asp.ped -d asp.dat --hardyWeinberg
At the bottom of the output, you should see a listing of marker genotype
statistics followed by two tables listing significant results for Hardy-Weinberg tests.
MARKER GENOTYPE STATISTICS
===========================
[Genotypes] [Founders] Hetero
MRK1 400 50.0% 0 0.0% 72.8%
MRK2 400 50.0% 0 0.0% 73.2%
MRK3 400 50.0% 0 0.0% 77.2%
MRK4 400 50.0% 0 0.0% 75.2%
MRK5 400 50.0% 0 0.0% 74.0%
MRK6 400 50.0% 0 0.0% 75.5%
MRK7 400 50.0% 0 0.0% 73.8%
MRK8 400 50.0% 0 0.0% 78.0%
MRK9 400 50.0% 0 0.0% 73.2%
MRK10 400 50.0% 0 0.0% 71.2%
MRK11 400 50.0% 0 0.0% 74.0%
MRK12 400 50.0% 0 0.0% 74.2%
MRK13 400 50.0% 0 0.0% 75.8%
MRK14 400 50.0% 0 0.0% 75.0%
MRK15 400 50.0% 0 0.0% 76.2%
MRK16 400 50.0% 0 0.0% 73.0%
MRK17 400 50.0% 0 0.0% 72.5%
MRK18 400 50.0% 0 0.0% 72.2%
MRK19 400 50.0% 0 0.0% 72.8%
MRK20 400 50.0% 0 0.0% 71.8%
Total 8000 50.0% 0 0.0% 74.1%
HARDY-WEINBERG CHECK AMONG ALL INDIVIDUALS
==========================================
N_HOM N_HET E_HET N_ALLELES ALLELES P-VALUE
MRK15 95 305 299 4 1-4 0.0333 A
MRK20 113 287 299 4 1-4 0.0249 A
Attempted Performed Failed [0.05] Failed [0.01]
Total Tests 20 20 2 0
HARDY-WEINBERG CHECK USING 200 UNRELATED INDIVIDUALS
====================================================
All 20 tested markers okay.
Attempted Performed Failed [0.05] Failed [0.01]
Total Tests 20 20 0 0
Each genotype table has the following 7 columns:
N_HOM column lists either the total number of homozygotes (for an asymptotic test) or the total number
of homozygotes followed by the number of minor allele homozygotes (for an exact test)
N_HET column lists the total number of heterozygotes
E_HET column lists the expected number of heterozygotes.
N_ALLELES column lists the total number of alleles for the marker
ALLELES column lists either the allele range (for the asymptotic test) or the
alleles used (exact test)
P_VALUE column lists the p-value for the test
TEST column indicates if an exact (E) or asymptotic (A) test was run
In the table "HARDY-WEINBERG CHECK AMONG ALL INDIVIDUALS", results for two significant tests are listed.
The first (Marker_15) is a microsatellite with 4 alleles in the range 1-4. If you look at the first few columns you
should also be able to verify that a total of 400 genotypes, consisting of 305 heterozygotes, and 95
homozygotes were available. The last column also indicates that PEDSTATS has performed an asympototic
X2 test (denoted by "A"), which gave a p-value of 0.033.
At the bottom of this table you'll find a brief summary of overall test results. Out of 20 tests attempted for the
asp.ped data set, PEDSTATS was able to complete 20 tests,
2/20 failed at the 0.05 level, and no test failed at the 0.01 level.
Summary mode for large data sets
The example data we've been using up to this point is quite simple. In practice, you'll probably be working with
data sets that are much more complicated -- perhaps with hundreds of individuals typed on thousands of markers. For
these very large data sets, PEDSTATS will switch to a summary mode for screen output and redirect detailed marker
information and (if applicable) Hardy-Weinberg results to a separate file. When this occurs, PEDSTATS indicates
that it is switching to summary mode, and produces a brief summary of marker quality.
Switching to summary output mode because there are more than 50 markers.
See file pedstats.markerinfo for detailed marker information and Hardy-Weinberg
test results.
If you look at the text file pedstats.markerinfo, you'll find the same detailed list of marker information we've discussed previously,
followed at the bottom of the file by two tables of Hardy-Weinberg test results.
HARDY-WEINBERG CHECK AMONG ALL INDIVIDUALS
==========================================
N_HOM N_HET E_HET N_ALLELES ALLELES P-VALUE
MRK15 95 305 299 4 1-4 0.0333 A
MRK20 113 287 299 4 1-4 0.0249 A
MRK23 193, 44 rare 207 186 2 1/2 0.0315 E
MRK26 255, 64 rare 145 179 2 1/2 0.0001 E
.
.
.
Attempted Performed Failed [0.05] Failed [0.01]
Total Tests 458 458 52 13
HARDY-WEINBERG CHECK USING 118 UNRELATED INDIVIDUALS
====================================================
N_HOM N_HET E_HET N_ALLELES ALLELES P-VALUE
MRK121 29 83 83 4, 5 pooled 132-149 0.0383 A
MRK276 36 79 75 3, 5 pooled 87-112 0.0080 A
MRK883 73, 24 rare 42 54 2, 5 pooled */120 0.0180 E
.
.
Attempted Performed Failed [0.05] Failed [0.01]
Total Tests 458 458 52 13
The second Hardy-Weinberg table will be a listing of significant tests using an unrelated sample. For these tests,
PEDSTATS first runs an
algorithm that selects a set of
Table 2 lists significant results for tests using 118 unrelated, genotyped individuals selected by PEDSTATS. If you look at the last table entry (MRK883), you may
notice some unfamiliar notation. In the column labelled "N_ALLELES" both the number of alleles used for the test (2) and
the number of alleles that were pooled (5) are listed. Since two allele groups are left after pooling, PEDSTATS performs an exact test - indicated
by an "E" in the last column. In the "ALLELES" column you'll find the allele groups used for the test. Here, allele 120 was tested against
the pooled 5-allele group (denoted by a *). The first two columns show the genotype distribution with respect to the pooled allele groups. Column 1 indicates that a total of 73 individuals were
homozygous; of these, 24 were homozygous for the minor (here, pool) allele. Column 2 indicates that there were 42 (pool/120) heterozygotes. In the third column labelled
"E_HET" you'll find the number of heterozygotes expected under the null hypothesis (54).



